recombination signal sequences
Highly conserved heptamer sequences, spacer sequences and conserved nonamer sequences that are adjacent to the V,D and J sequences in the heavy chain region of DNA and the V & J sequences in the light chain DNA region.
Used during V(D)J recombination by recombination activating gene to align the gene segments.
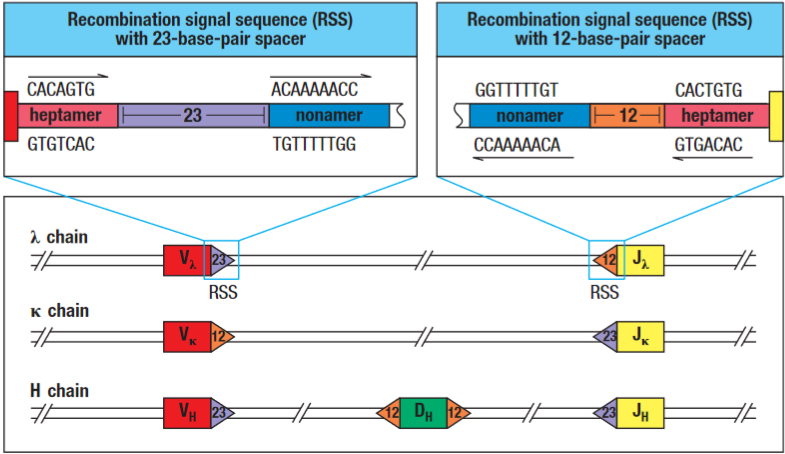 The heptamer - 5’CACAGTG3’ which is continuous with the coding sequence → spacer region (12 or 23 bp) → nonamer 5’ACAAAAACC3’.
The heptamer - 5’CACAGTG3’ which is continuous with the coding sequence → spacer region (12 or 23 bp) → nonamer 5’ACAAAAACC3’.
These sequences are the consensus sequences but can vary from one gene segment to another, even in the same individual, as there is some flexibility in the recognition of these sequences by the enzymes that carry out the recombination (recombination activating gene).
Tip
A gene segment flanked by an RSS with a 12-bp spacer typically can be joined only to one flanked by a 23-bp spacer RSS. This is known as the 12/23 rule. V and J domains on the λ chain have the inverse spacer orientation to κ to prevent cross binding. Additionally Vh and Jh cannot be joined directly without the Dh gene segment due to the 23 bp spacers on each of them.
Infrequently D-D joining can occur (violating the 12/23 rule) and is found in 5% of human antibodies and is the major mechanism accounting for the unusually long CDR3 loops found in some heavy chains.
2023-10-17T12:04
References
https://en.wikipedia.org/wiki/Recombination_signal_sequences { Janeway’s Immunobiology, 9th edition Chapter 5-4