V(D)J recombination
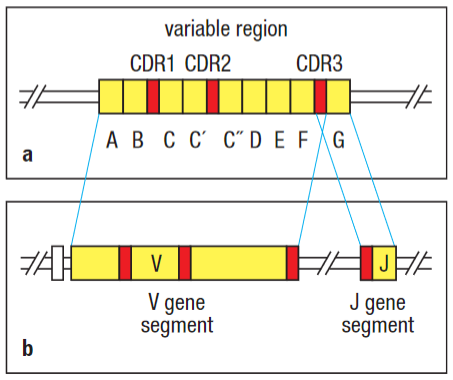
As the b-cell develops in the bone marrow the first 95-101 AA of the variable region originate from the V gene segment. V gene makes up part of CDR3, and the remainder originate from a J gene segment.
in nonlymphoid cells, the V-region gene segments remain in their original germline configuration.
VL is encoded by V and J genes and VH is encoded in three segments (V, J and D genes).
In the heavy chain, the DH gene segment is joined to a JH then the VH gene segment rearranges the DJH to make a complete VH-region exon. As with the light-chain genes, RNA splicing joins the assembled V-region sequence to the neighbouring C-region gene. This is known as V(D)J recombination.
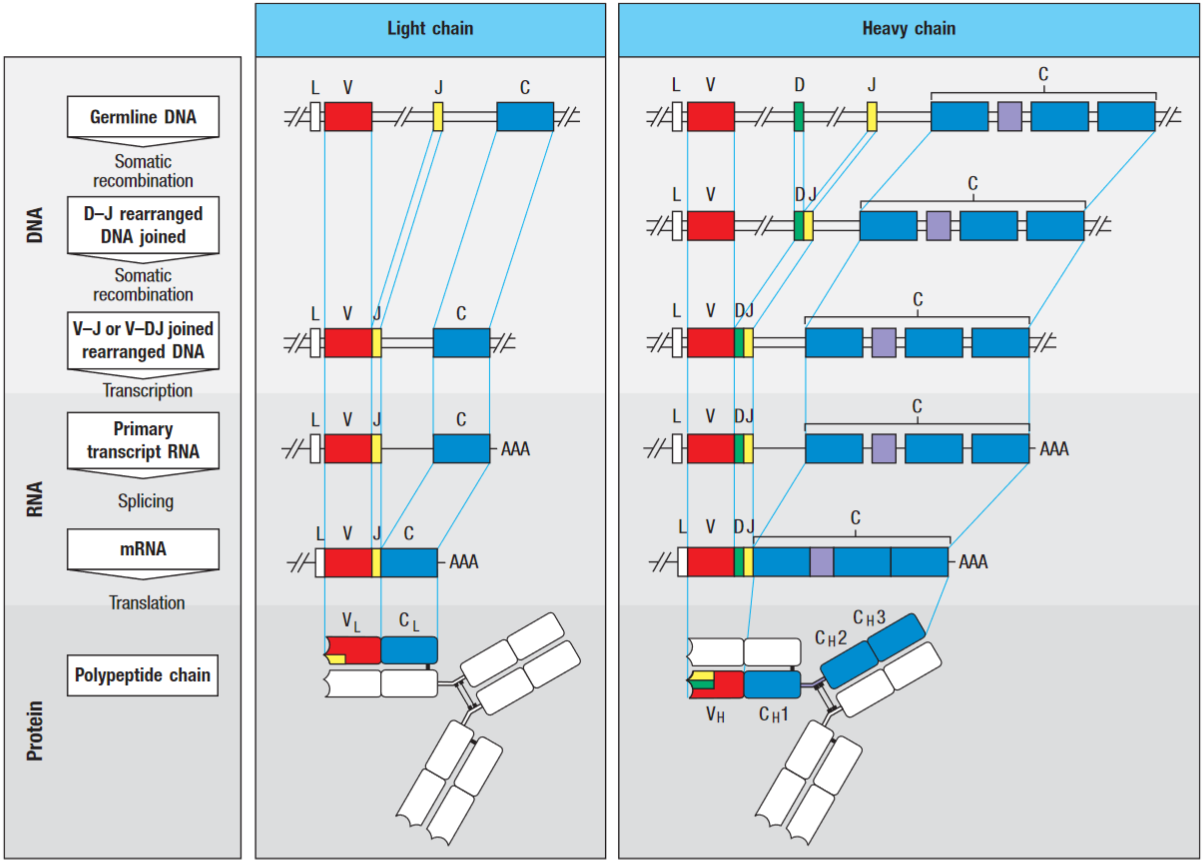
The V-gene segment is preceded by an exon encoding a leader peptide (L), which directs the protein into the cell’s secretory pathways, and then cleaved.
There are multiple copies of V, D and J gene segments in the germline DNA. The random selection of just one gene segment of each type produces the diversity of variable regions among immunoglobulins.
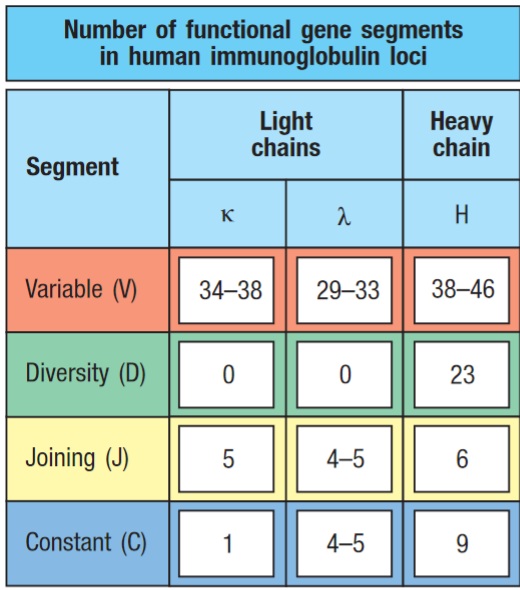 Number of functional gene segments for the V regions of human heavy and light chains. As there are many V, D and J gene segments in germline DNA, no single gene segment is essential. This has lead to the formation of a large number of pseudogenes.
Number of functional gene segments for the V regions of human heavy and light chains. As there are many V, D and J gene segments in germline DNA, no single gene segment is essential. This has lead to the formation of a large number of pseudogenes.
The gene segments are organised in three genetic loci - the κ, λ and heavy-chain loci - each of which can assemble a complete V-region sequence.
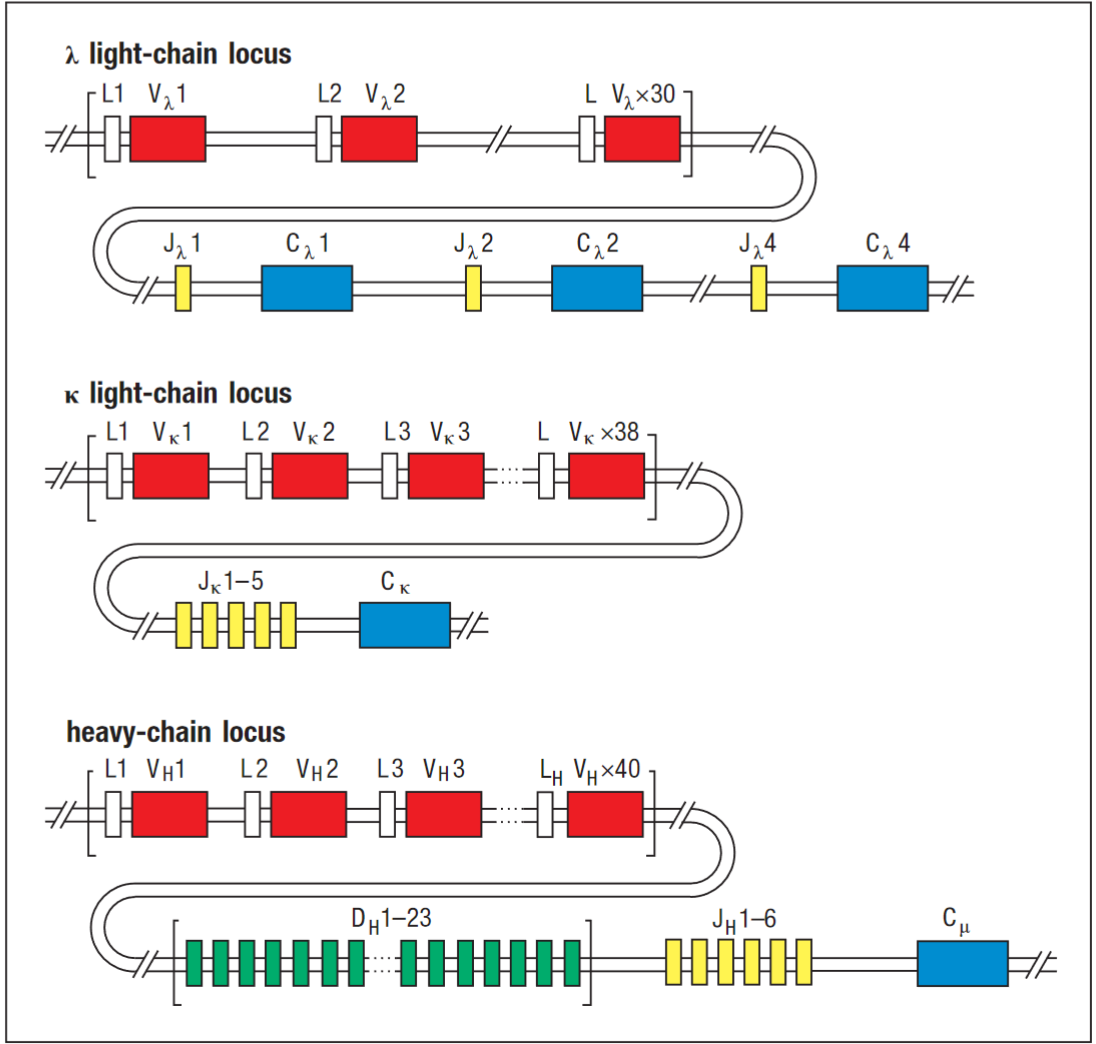
Rearrangement of V, D and J gene segments is guided by flanking DNA sequences
To ensure rearrangements are regulated they are guided by conserved noncoding DNA sequences, called recombination signal sequences (n.b especially the 23 rule).
The mechanism of DNA rearrangement is similar for heavy & light chain loci (HC has additional recombination event).
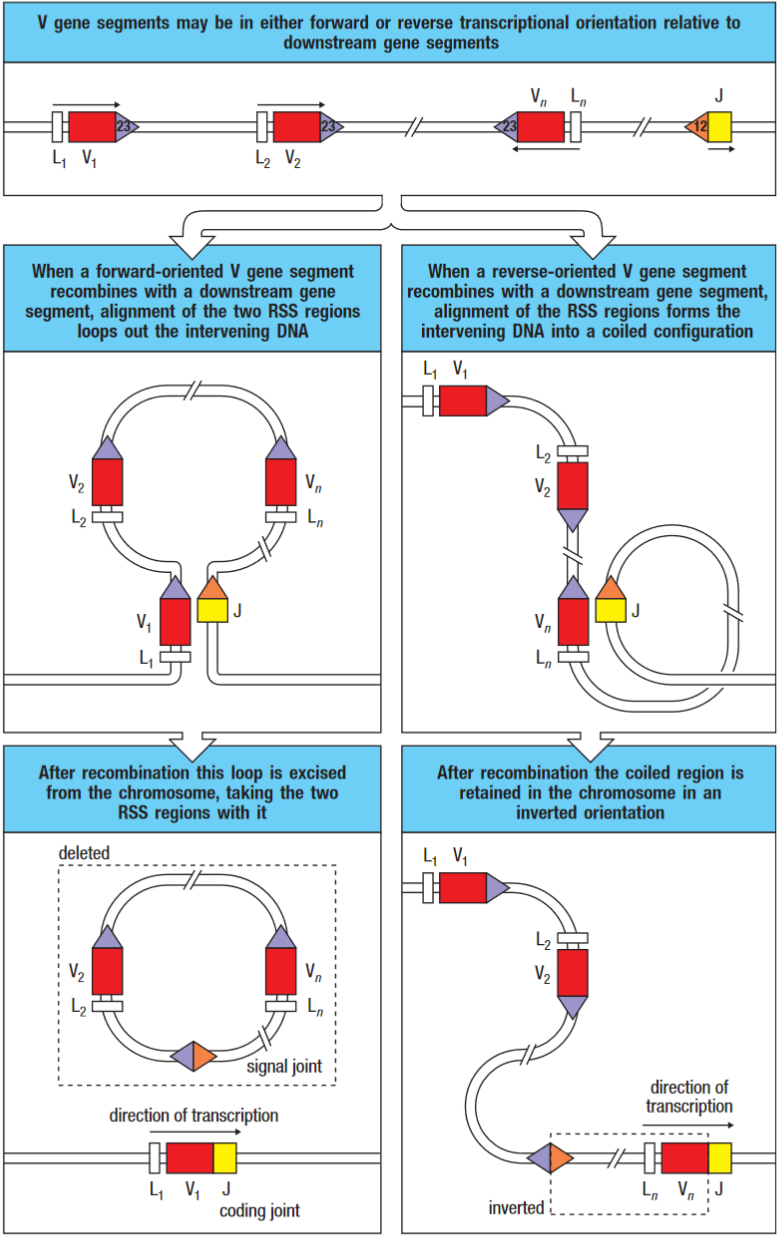 *Standard rearrangement on the left. Rearrangement by inversion is shown on the right.
*Standard rearrangement on the left. Rearrangement by inversion is shown on the right.
Two RSSs are bought together by interactions between proteins that recognise the length of the spacers and enforce the 23 rule for recombination. The DNA is cleaved by endonuclease activity at two locations and is then re-joined with the heptamer sequences head-to-head, forming a signal joint. When the joining segments are in the same orientation, the signal joint is contained in a circular piece of extrachromosomal DNA which is lost from the genome when the cell divides. The V and J gene segments, which remain on the chromosome, join to form the coding joint.
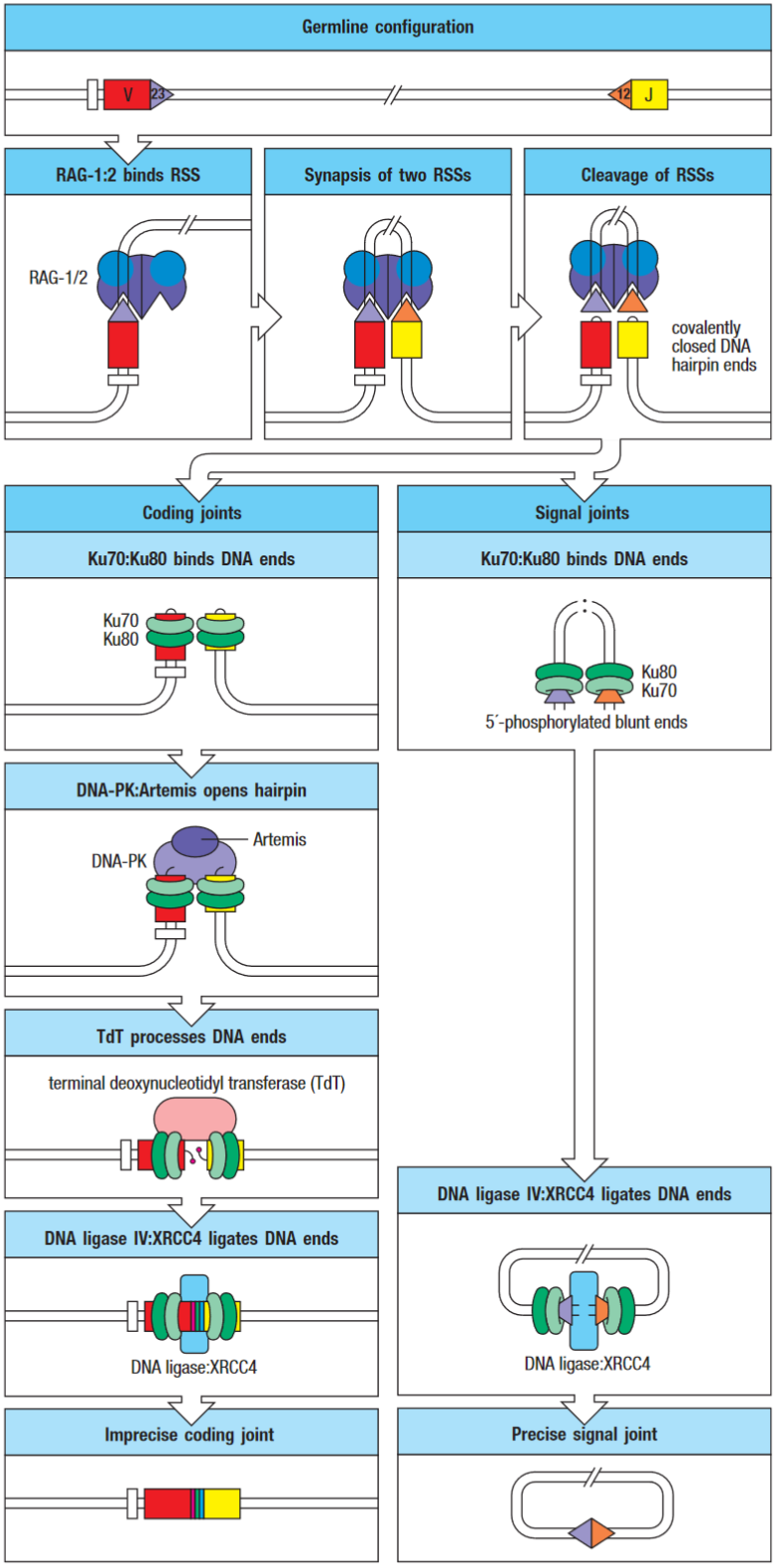
V(D)J recombinase carry out the somatic recombination. The lymphoid-specific components of the recombinase are called RAG-1 and -2. These genes are expressed in developing lymphocytes only while they are engaged in assembling their antigen receptors.
Tip
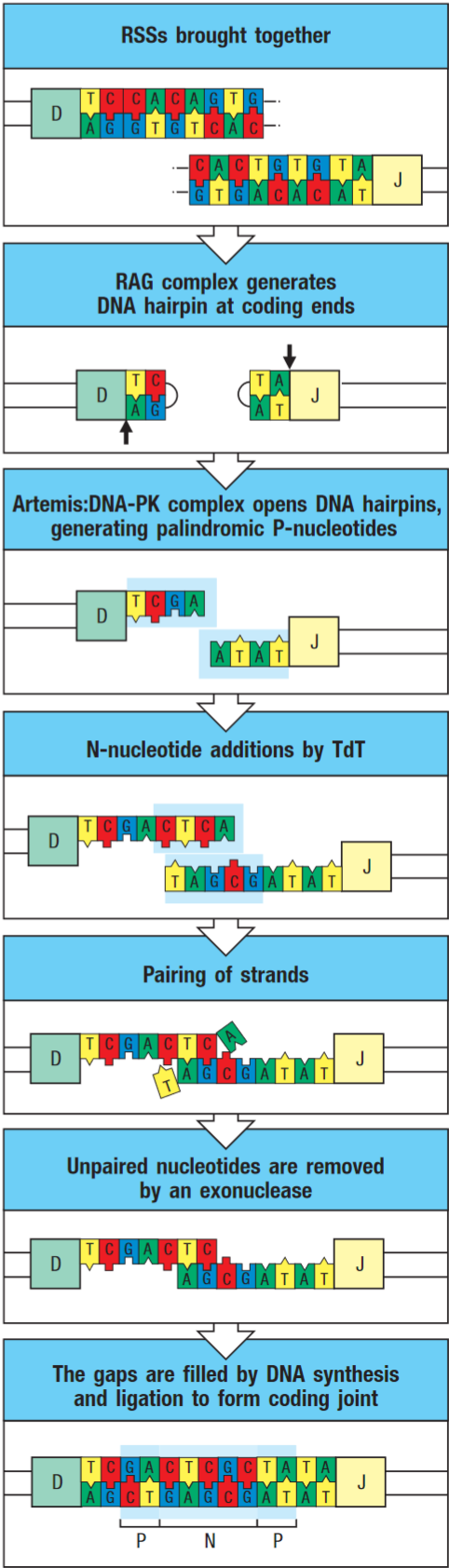
DNA-PK:Artemis complex opens the hairpin at a random site yielding either two flush-ended DNA strands or a single-strand extension. The cut end is then modified by terminal deoxynucleotidyl transferase (TdT) and exonuclease, which randomly add and remove nucleotides respectively.
Therefore the coding joint is imprecise, this variability is known as junctional diversity.
TCR V(D)J recombination
The TCR gene segments rearrange during t-cell development to form complete V-domain exons. TCR gene arrangement takes place in thymus; the mechanics are similar to antibodies as explained above.
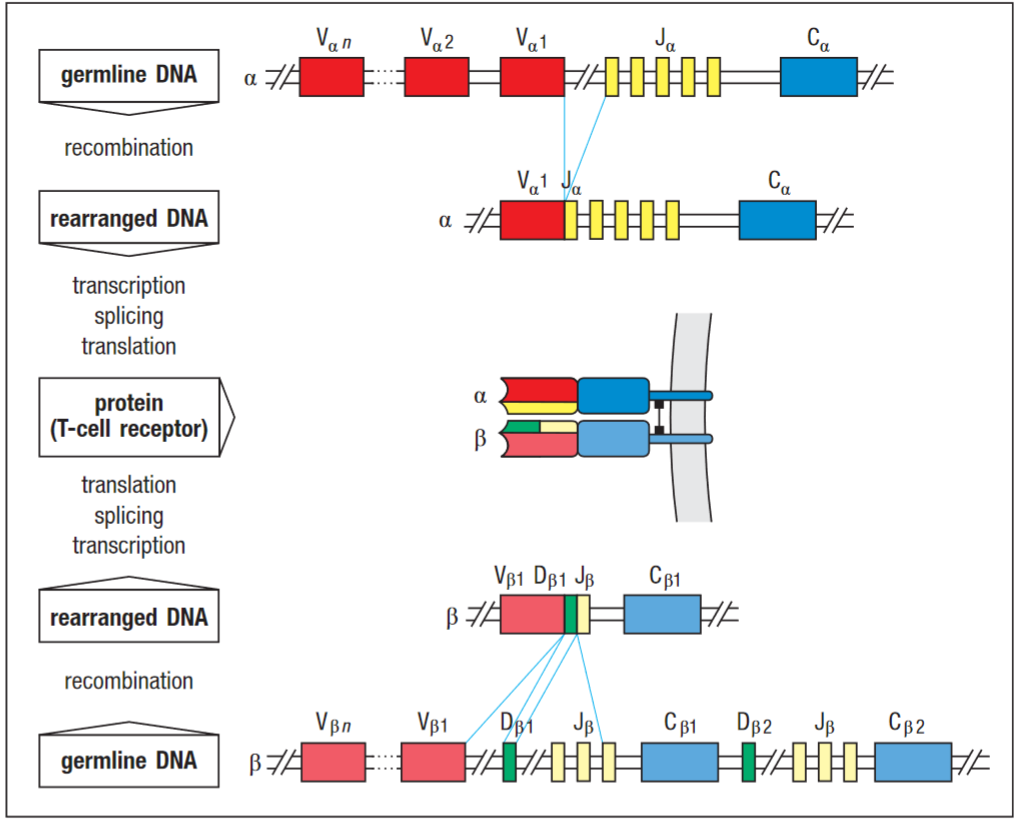
C-regions of TCR are much simpler than those of the immunoglobulin heavy-chain locus.
Abstract
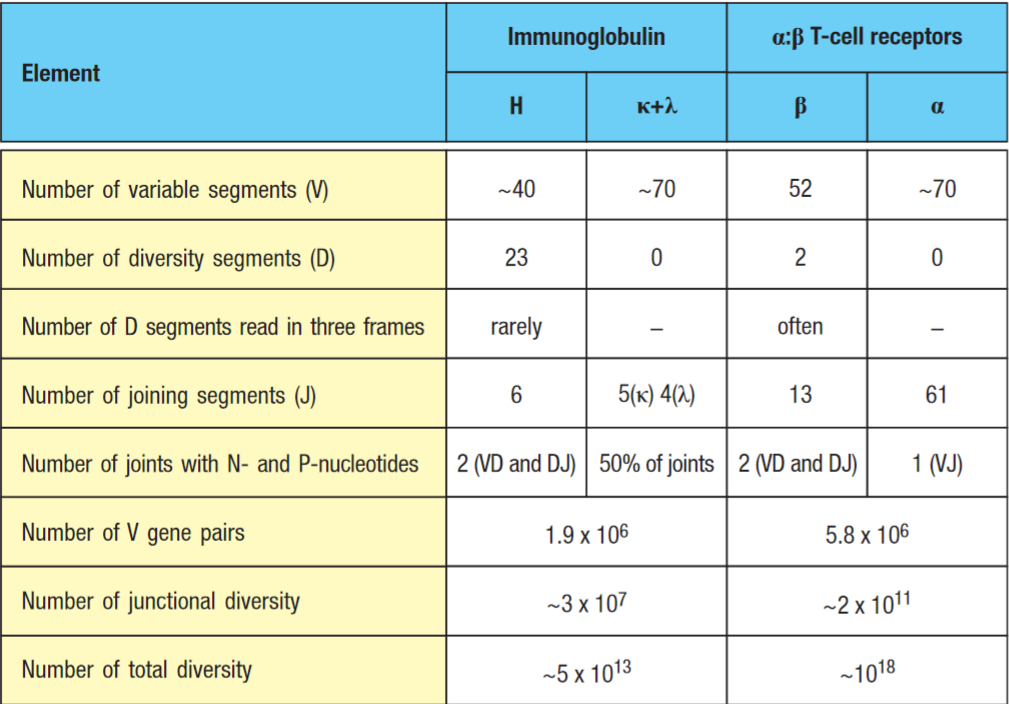
TCR variability is concentrated in the CDR3 region as it contacts the unique peptide component of the MHC. Most variability is in the junctional regions, see junctional diversity, which are encoded by V,D and J gene segments and modified by P- and N-nucleotides. The TCRα locus contains many more J gene segments than either of the immunoglobulin light chain loci. (humans: 61 Jα gene segments vs 5 J lc segments). Thus, most of the diversity resides in the CDR3 loops that contain the junctional region and form the centre of the antigen-binding site.
2023-10-17T09:59
References
{ Janeway’s Immunobiology, 9th edition Chapter 5